Introduction
It looks like I have made a mistake when zipping the dMRI files (DWI_*_TC_std.nii.gz).
I plan to fix that as soon as I have time. Meanwhile, you should be able to extract the readable nifti files DWI_3000_TC_std.nii via
"tar --transform='s:.*/::' -xzf DWI_3000_TC_std.nii.gz".
The BMCR is a resource that provides access to anterograde and retrograde neuronal tracer data, currently restricted to injections into the prefrontal cortex of a marmoset brain.
We are currently extending the resources with ISH gene expression image data from the Marmoset Gene Atlas
(https://gene-atlas.brainminds.jp/),
and integrating data from anterograde tracer injections covering the entire marmoset cortex. It comprises post-processed data and tools for exploring and reviewing the data, and for integration with other marmoset brain image databases and cross-species comparisons.
We visualize data of different individuals in a common reference space at an unprecedented high resolution. Major features are:
- Image data of anterograde and retrograde neuronal tracers.
- The BMCR-Explorer for reviewing the data in high resolution
- dMRI tractography data that is associated with anterograde injection sites.
- Transformation fields to map between other marmoset brain image databases and atlas (Brain/MINDS Marmoset Reference Atlas, Marmoset Brain Mapping, Marmoset Brain Connectivity Atlas).
- Transformation fields to the human MNI space
- Flatmap stacks of neural tracer data (Including the all neccesary data to map other image data to flatmap stacks).
- Nora-StackApp for reviewing the data
- A left-right symmetric population-average brain image template based on serial two-photon tomography along with a left-right symmetric population-average HARDI, Nissl, and backlight image.
- We integrated retrograde neural tracer data from the Marmoset Brain Connectivity Atlas project.
Coming next
- ISH image data in nifti format.
- High resolution data from anterograde tracer injections covering the entire corext.
News
- 2024/10: Integrated image data from 260 Genes of adult marmosets from the Marmoset Gene Atlas (https://gene-atlas.brainminds.jp/) into the BMCR-Explorer
- 2024/9: BMCR-Explorer: Anterograde tracer segmentations have been updated (better coverage at brain boundaries)
Table of Contents
Major Publications
These are our related publications. Please note that the data also incooperates data from other sources as well. Please carefully read the license files or hints in the BMCR-Explorer.
STPT tracer data integration and data acquision
-
A. Watakabe, T, Tani, H. Abe, H. Skibbe, N. Ichinohe, T. Yamamori
In bioRxiv, 2024
-
A. Watakabe, H. Skibbe, K. Nakae, H. Abe, N. Ichinohe, M. F. Rachmadi, J. Wang, M. Takaji, H. Mizukami, A. Woodward, R. Gong, J. Hata, D. C. Van Essen, H. Okano, S. Ishii, T. Yamamori
In Neuron, 2023
-
H. Skibbe, M.F. Rachmadi, K. Nakae, C. E. Gutierrez, J. Hata, H. Tsukada, C. Poon, K. Doya, P. Majka, M. G. P. Rosa, M. Schlachter, H. Okano, T. Yamamori, S. Ishii, M. Reisert, A. Watakabe.
In PLoS Biology, 2023
ISH data integration
-
C. Poon, M.F. Rachmadi, M. Byra, M. Schlachter, B. Xu, T. Shimogori, H. Skibbe
In IEEE International Symposium on Biomedical Imaging Proceedings, 2023
Video: The video above illustrates some of the major parts of our image processing pipeline. It is a bit outdated. By that time, we completed the image processing for 41 cortical injections.
Brain/MINDS Marmoset Connectivity Resource
BMCR-Explorer
The BMCR-Explorer is publicly available online.
You might find our introduction helpful.
Or directly start by clicking on one of the example images below.
The BMCR-Explorer works best with google-chrome, but should also work with the chromium, firefox and edge browsers.
Safari might work. It also has touch support. It won't work with the Microsoft Internet Explorer :-(. We plan to release a downloadable offline version of the BMCR-Explorer in the future.
Click here or on one of the examples below to open the BMCR-Explorer
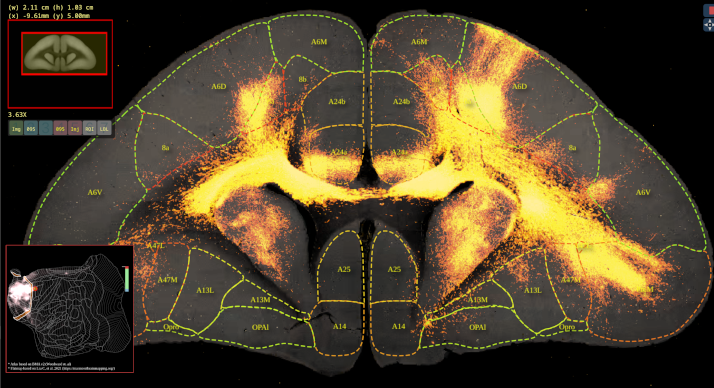
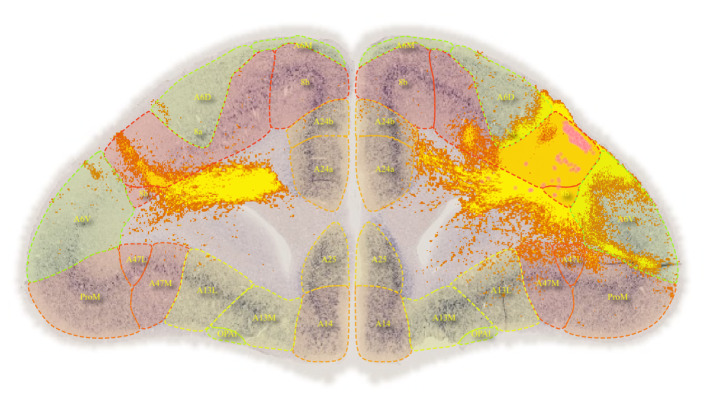
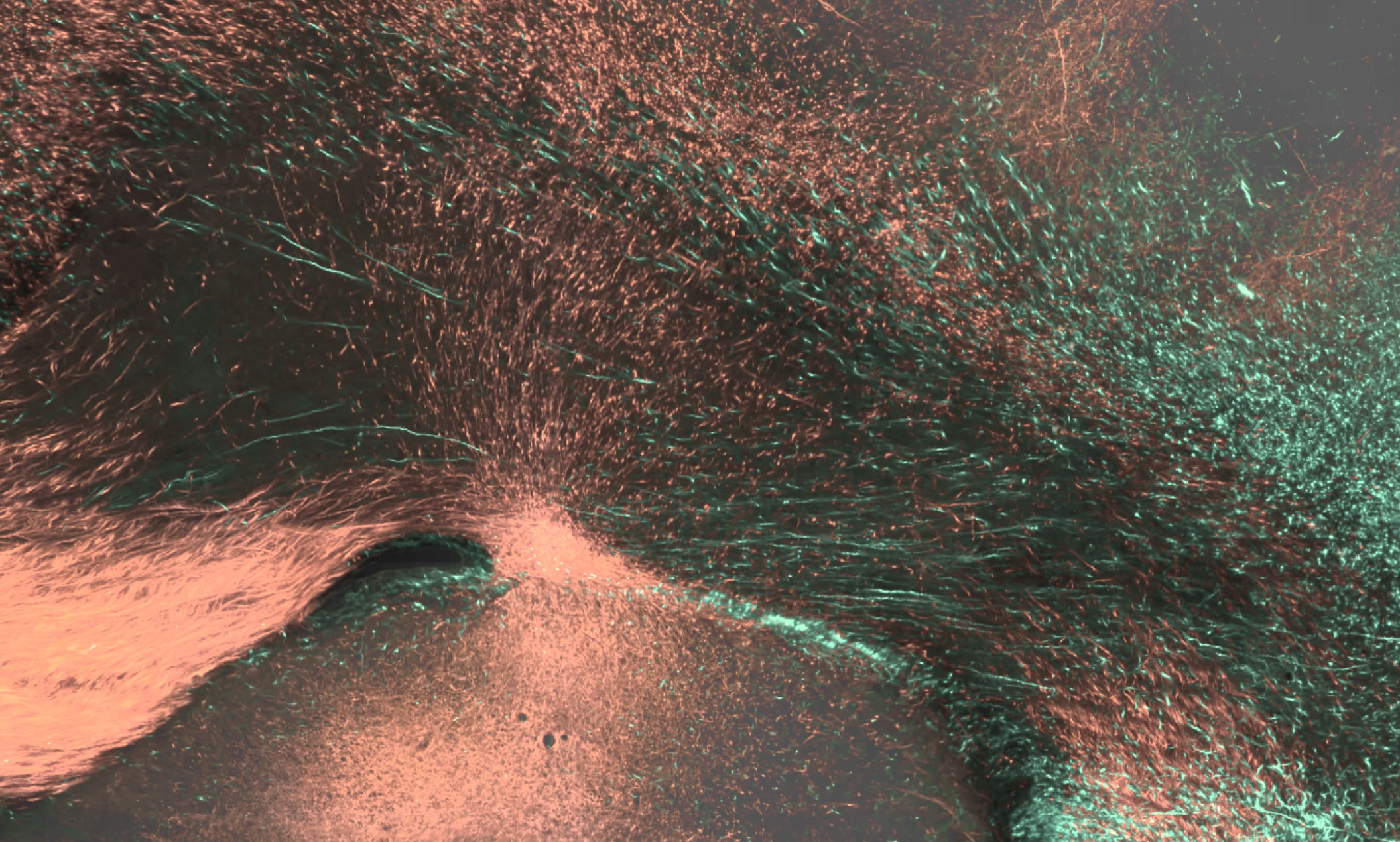
BMCR-Explorer Example 1: An anterograde tracer segmentation mask on top of the original serial two-photon tomography image (click on the figure to open the image in the BMCR-Explorer).
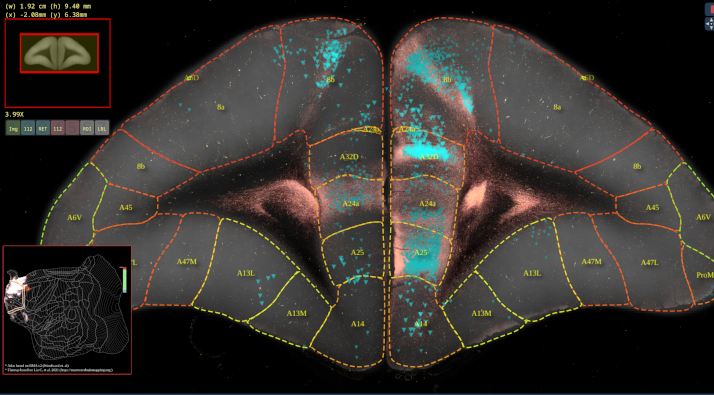
BMCR-Explorer Example 2: An anterograde and retrograde tracer (click on the figure to open the image in the BMCR-Explorer).
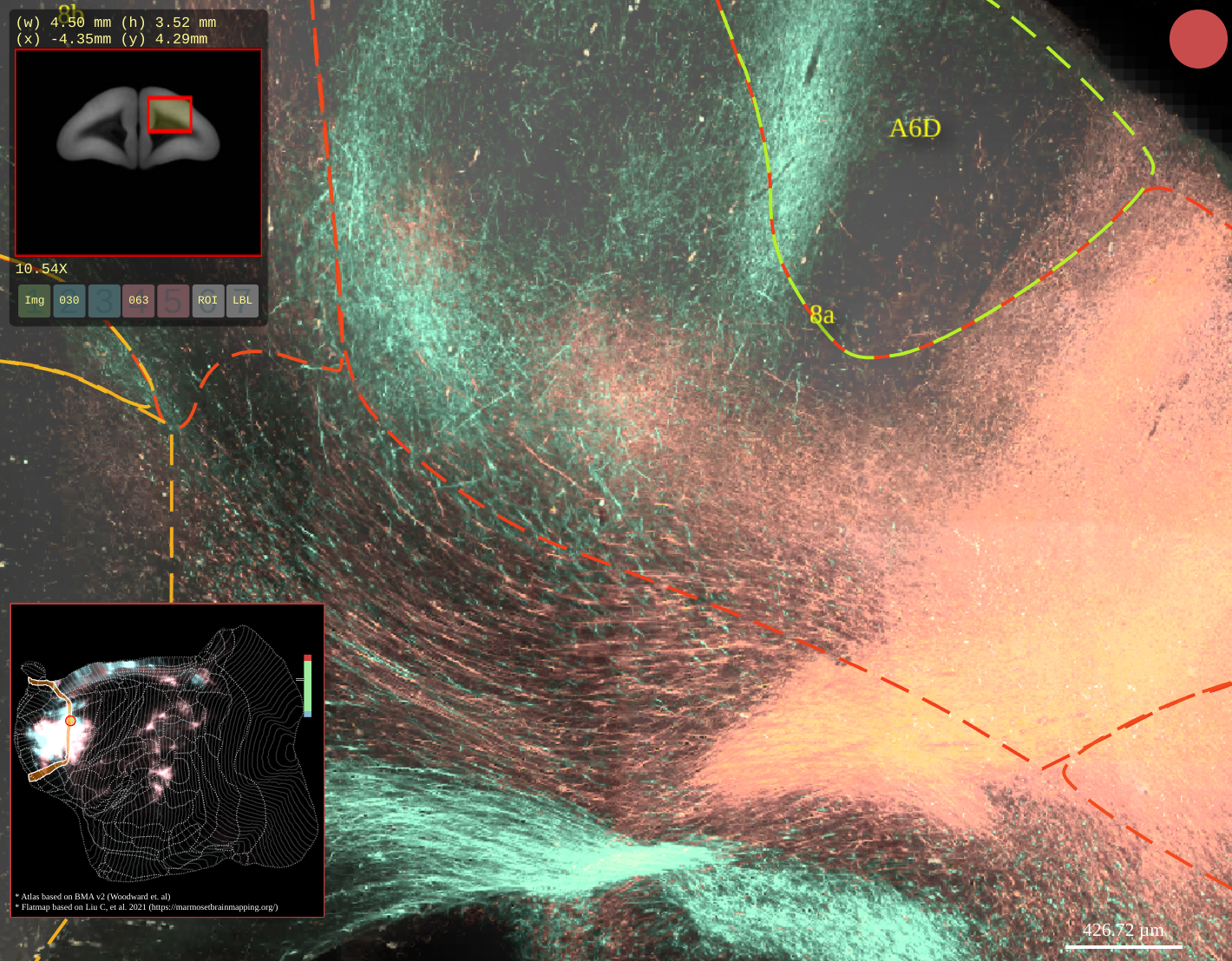
BMCR-Explorer Example 3: Two serial two-photon tomography images of anterograde neural tracers from two different marmoset brains (click on the figure to open the image in the BMCA-Explorer)..
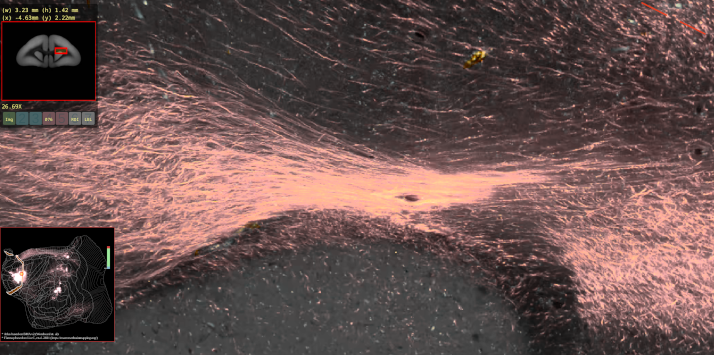
BMCR-Explorer Example 4: A serial two-photon tomography image of an anterograde neural tracer in the marmoset brain (click on the figure to open the image in the BMCR-Explorer).
BMCR-Explorer Example 5: Segmentation of anterograde neural tracer signal on top of an ISH image (click on the figure to open the image in the BMCR-Explorer).
Nora-StackApp and Data Download and License
Nora-StackApp and data in the STPT reference image space
Video: Demonstration of the Nora-StackApp. The Nora-StackApp based on the
nora medical imaging platform, which we extended with functions so that it interacts with BMCR-Explorer and flatmap stacks.
License
You can download the data from here. Further instructions for installing the Nora-stack app can be found here: Nora-Howto
Our image data is published under the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/deed.en).
The BMCR-Explorer and the downloadable dataset also includes data derived from four atlases that are work of other groups, which you might find helpful in reproducing and exploring our findings. Please carefully read the accompanying ‘Readme’ files, as some of the data is released under different licenses. If you find these data useful and plan to use it, then you must cite the original work, and accept their licenses. A summary of the atlas licenses is provided below:
(1) Brain/MINDS 3D Marmoset Reference Brain Atlas 2019 (abbreviated as BMA)
(2) Data from the Marmoset Brain Connectivity Atlas (abbreviated as MBCA)
(3) Atlas data from the marmoset brain mapping project (version 2+3) (abbreviated as MBM)
-
Nora-StackApp
The Nora-StackApp is based on the Nora medical imaging platform. Therefore, you might find the
First Steps tutorial useful, too.
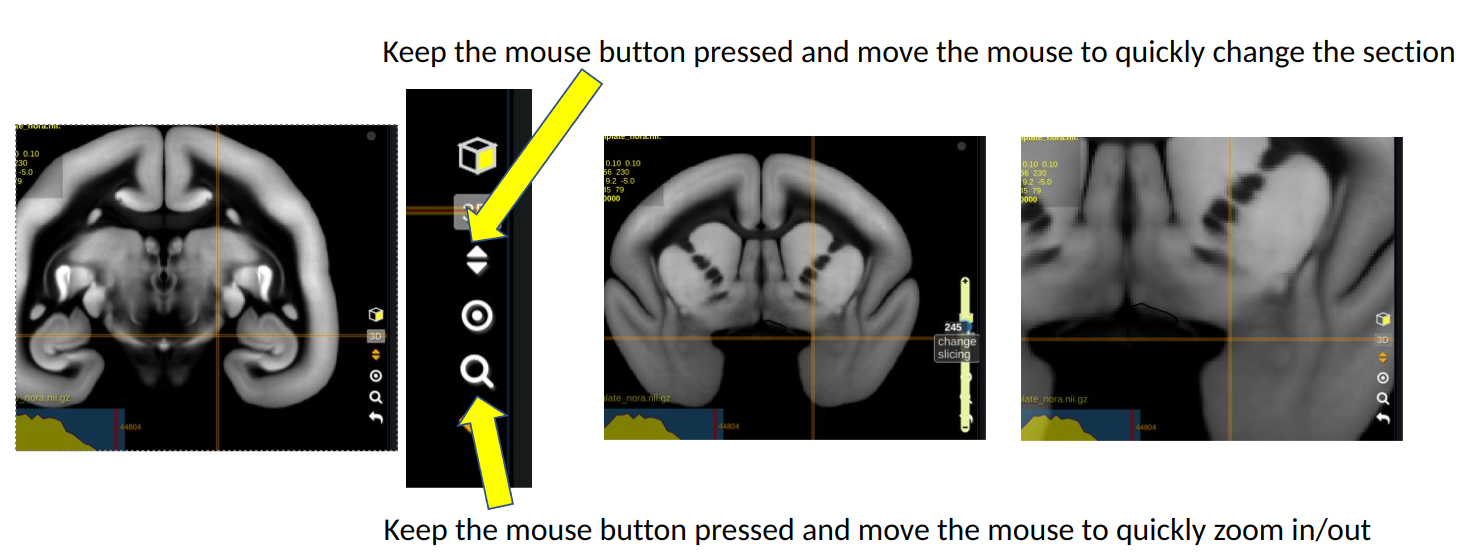
Basic keyboard and mouse controls:
Explained in Tutorial 00
- Keep the left mouse button pressed while moving the cursor to navigate withing slices
- Keep the right mouse button pressed while moving the cursor to shift a section
- Use the mouse wheel to switch slices
- Use the mouse wheel while keeping the control key pressed to zoom
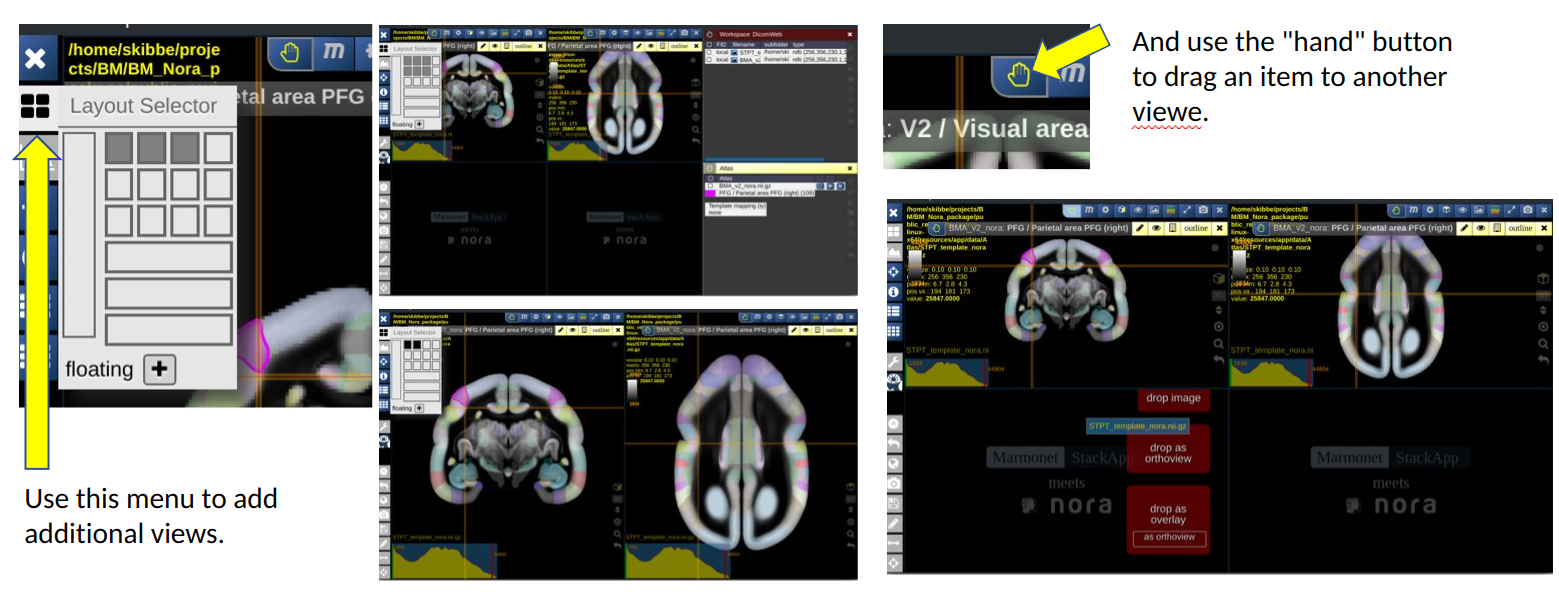
You can also use the quick navigation menu as shown below:
Installation
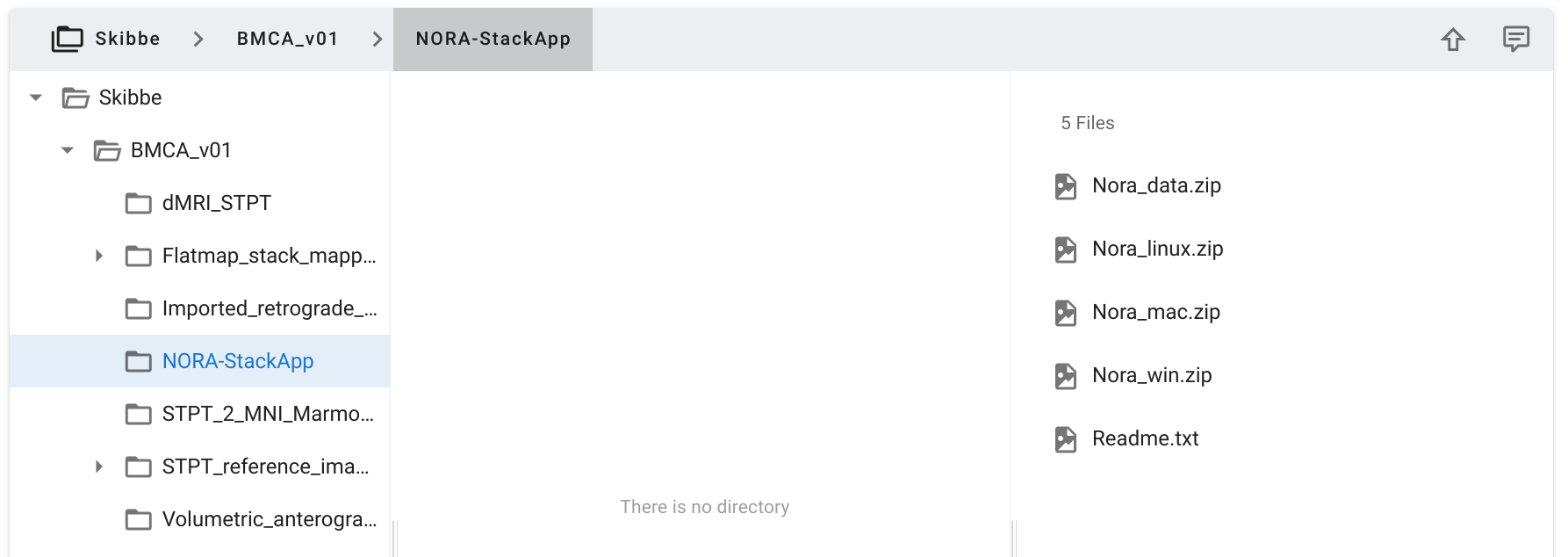
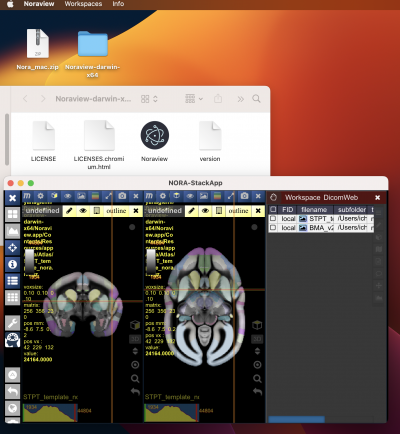
Download a copy from the
RIKEN CBS data portal. Select the NORA-StackApp folder as shown in the image below.
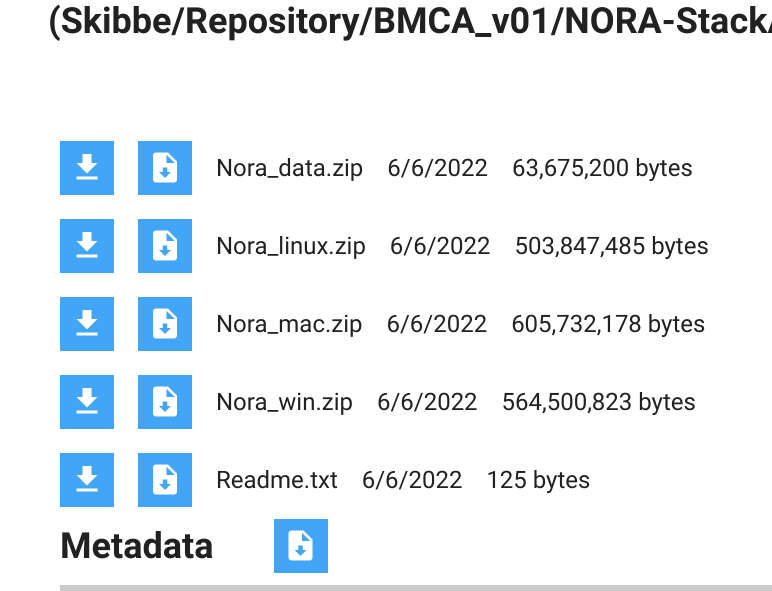
To download the StackApp, click on the

symbol in the upper right corner. It will open the download window (you might need to allow pop-up windows for the repository site). Below a screenshot of the download window:
Please download the version for your OS (linux,mac or win). We have tested it on Ubuntu Linux, Windows 11 and an M1 Mac. Below a screenshot that illustrates how to run it from a Mac. Windows and Linux work in a similar way. You won't need to install the app, just unzip the folder. In the example below, we used the Desktop as destination. In a sub-folder of the unzipped folder, you will find the “Noraview” application (on Windows Noraview.exe). Starting it should bring up the main interface as shown below:
The work-spaces menu provides several pre-defined work-spaces for marmosets.
Download Tracer Data
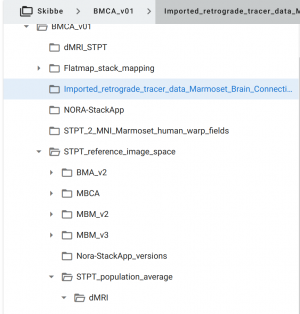
When you download the Nora-StackApp, you already have the atlases and the template images that are used for the work-spaces. But you might want to download tracer data image stacks and additional atlases from the RIKEN CBS data repository:
- Anterograde tracer data: “Volumetric_anterograde_tracer_data”
- Retrograde tracer from the Marmoset Brain Connectivity Project
- Atlases: Use the Nora-optimized versions under “STPT_reference_image_space” → “Nora-StackApp_versions”
Marmoset Workspaces
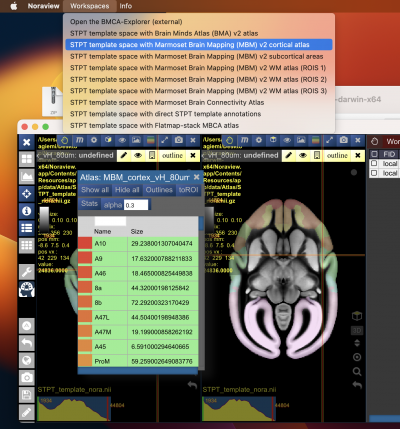
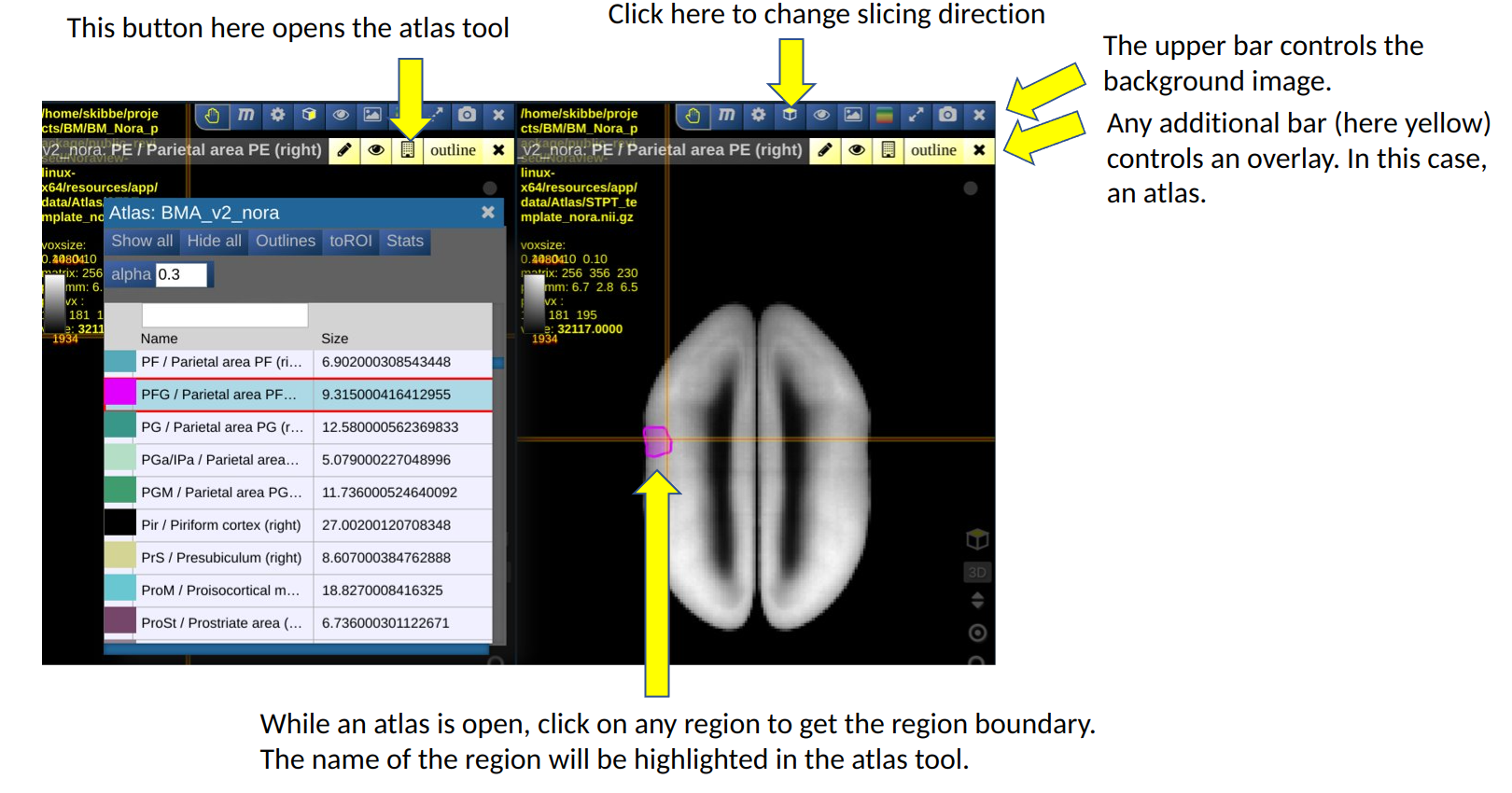
The Nora-StackApp shows multiple 3D image stacks in a grid of views. Each view can show a section of an image. A view shows one main image (e.g. a background image) and can have multiple “overlays”. An overlay can be another image, such as a tracer signal, or an atlas. The example below shows the default marmoset workspace with two views, each showing a different slicing. It also shows the atlas tool.
You can increase or decrease the number of views. Having more views can be helpful to show two different images side-by-side. Reducing the number of views gives a view more space on the screen.
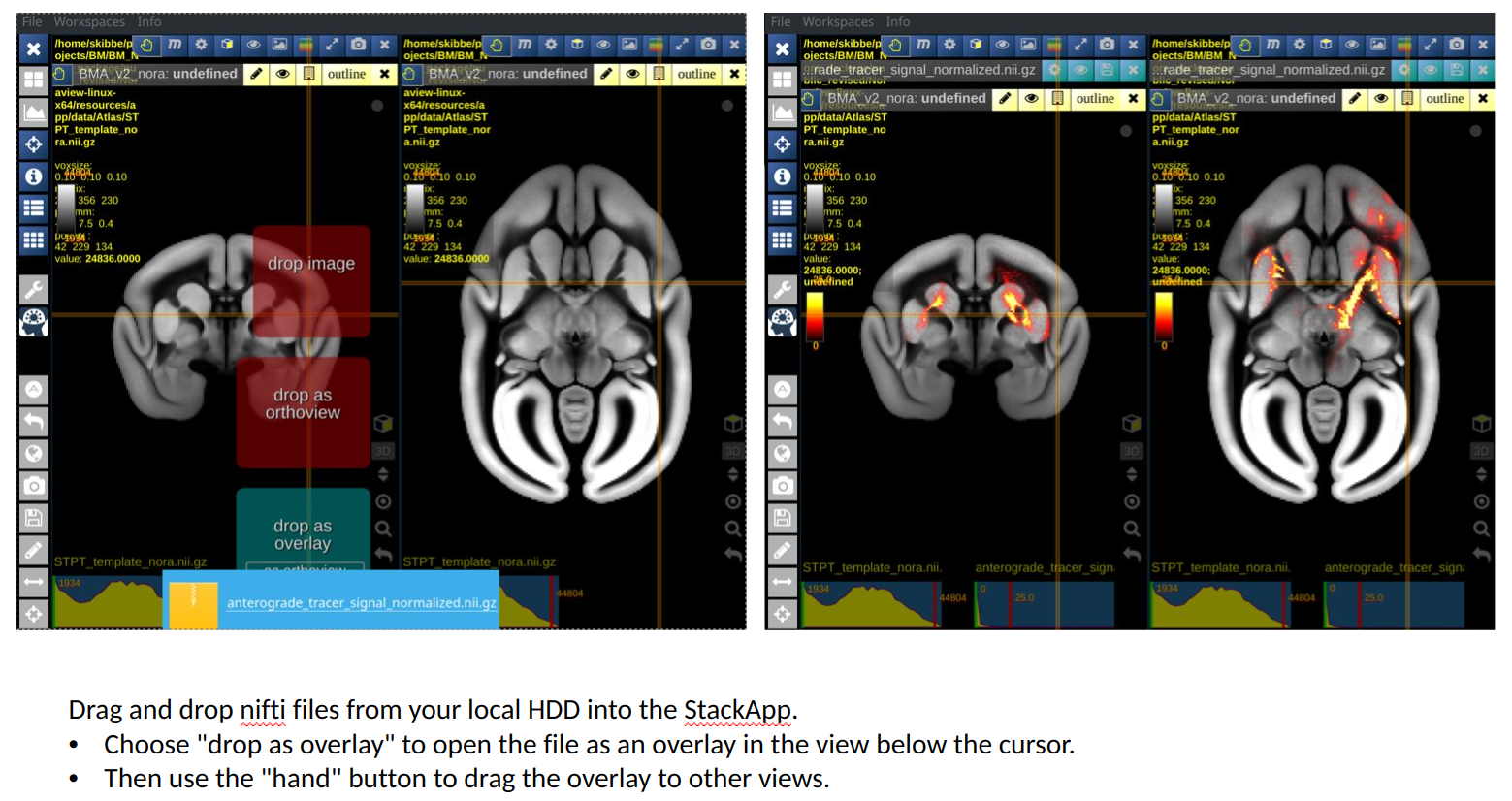
The image below shows the default workspace, that shows our average template as background image. We have selected a nifti file with an anterograde tracer signal from our hard disk, and use the drag-and-drop feature of Nora to drop it into the first view as a new overlay. We then took the hand button of the tracer overlay and dropped it into the second view as well.
Open the BMCA-Explorer, and use the brain-symbol shown below, and drag and drop it into the explorer to set the current Explorer position to the same position as in Nor
The video below shows a demonstration, including interaction the with BMCA-Explorer:
Flatmap Stack and Human Brain Space Mapping
The BMCR dataset includes the STPT template images (with a population average STPT background image, a population average Nissl image, a population average backlit image and a population average HARDI. It further incooperates measurements derived from the population average HARDI. It also includes
the ANTs warping field that maps data from our STPT template space to a flatmap-stack, and ANTs warping fields to map between other marmoset brain atlases (MARMOSET BRAIN CONNECTIVITY ATLAS, MARMOSET BRAIN MAPPING version 2 and 3, Brain/MINDS Marmoset Reference Atlas v1.1.0). It also includes the warping field between the STPT image space and the human MNI space.
Video: Demonstration of the warping field mapping from the STPT template image space to a human brain
Howto Flatmap-stack
Some code examples showing how to use the ANTs image registration toolkit to map marmoset brain images to a flatmap stack can be found here: https://github.com/BrainImageAnalysis/flatmap_stack/wiki.
Video: Demonstration of the warping field mapping from the STPT template image space to the 3D flatmap stack